File size: 5,630 Bytes
0087201 bf3d8d2 de88378 bf3d8d2 ac75658 bf3d8d2 ac75658 bf3d8d2 1a61527 bf3d8d2 de88378 bf3d8d2 1a61527 bf3d8d2 de88378 bf3d8d2 ac75658 bf3d8d2 ac75658 0bcc800 ac75658 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 |
---
license: mit
datasets:
- TongjiZhanglab/NuCorpus-15M
---
# NuSPIRe: Nuclear Morphology focused Self-supervised Pretrained model for Image Representations
## Model description
NuSPIRe (Nuclear Morphology focused Self-supervised Pretrained model for Image Representations) is a deep learning model designed to extract nuclear morphological features from DAPI-stained images. The model utilizes self-supervised pretraining, learning from 15.52 million unlabeled nuclear images from diverse tissues. NuSPIRe is optimized for biomedical image analysis tasks such as cell type identification, perturbation detection, and gene expression prediction, particularly excelling in scenarios with limited annotations.
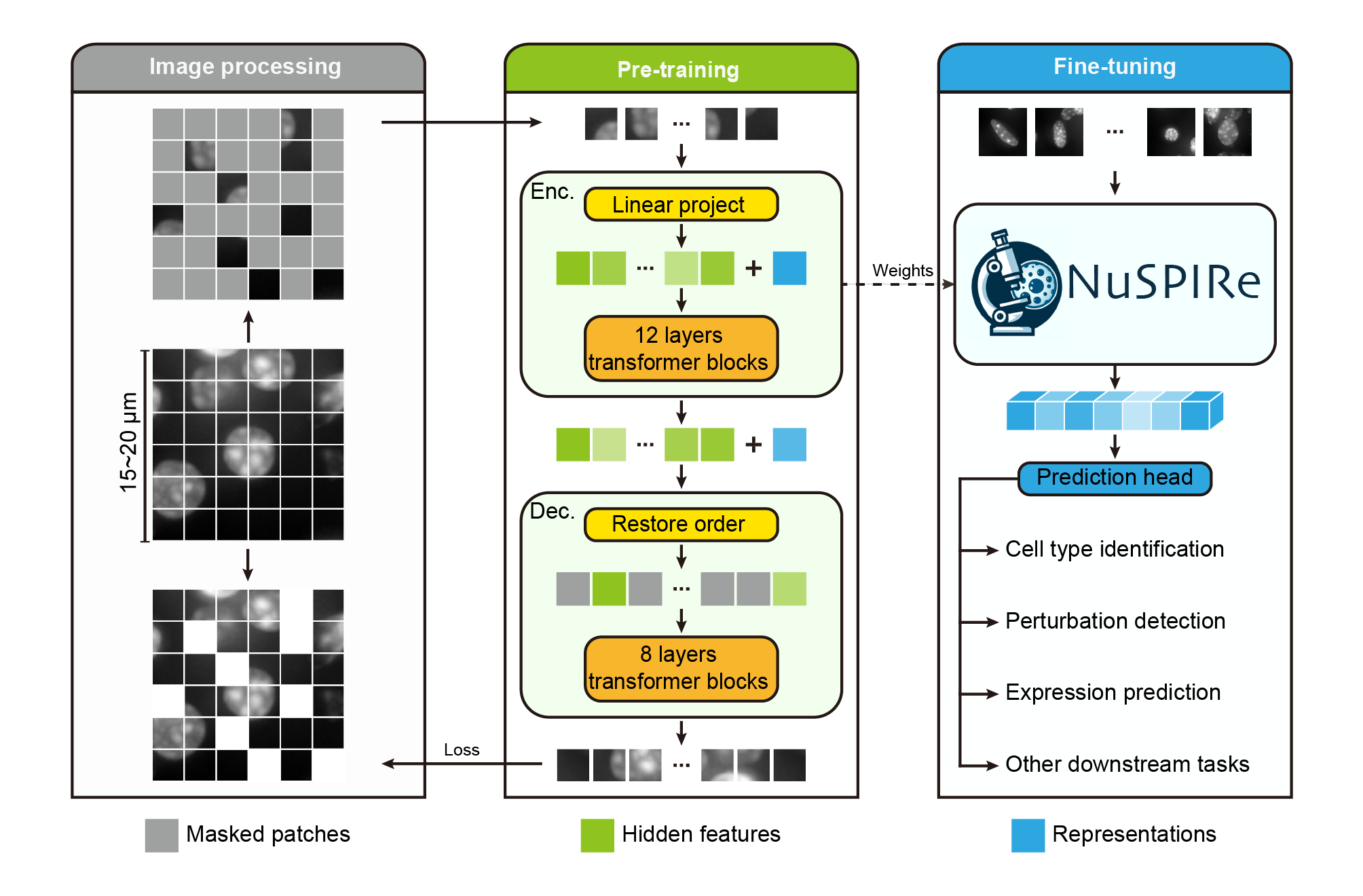
## Training Details
- **Pretraining Dataset**: NuCorpus-15M, a dataset comprising 15.52 million cell nucleus images from both human and mouse tissues, spanning 15 different organs or tissue types.
- **Input**: The model processes DAPI-stained images of cell nuclei, which are commonly used to visualize nuclear structure.
- **Tasks**: NuSPIRe is capable of handling various downstream tasks, including cell type identification, perturbation detection, and predicting gene expression levels.
- **Framework**: The model is implemented in PyTorch and is available for fine-tuning on specific tasks.
- **Pre-training Strategy**: NuSPIRe was trained using a Masked Image Modeling (MIM) approach for self-supervised learning, allowing it to extract meaningful features from nuclear morphology without needing labeled data.
- **Downstream Performance**: The model significantly outperforms traditional methods in few-shot learning tasks and performs robustly even with very small amounts of labeled data.
## Usage
### Representation Extraction
To extract representations using the pre-trained NuSPIRe model, please refer to the code example below:
```python
# Import necessary libraries
import torch
import requests
from PIL import Image
import lightning.pytorch as pl
from transformers import ViTModel
from torchvision import transforms
# Set random seed for reproducibility
pl.seed_everything(0, workers=True)
# Open an example image
url = 'https://huggingface.co/TongjiZhanglab/NuSPIRe/resolve/main/Images/image_aabhacci-1.png'
image = Image.open(requests.get(url, stream=True).raw).convert('L')
# Define the image preprocessing transformations
transform = transforms.Compose([
transforms.Resize((112, 112)), # Resize image to 112x112 pixels
transforms.ToTensor(), # Convert PIL image to PyTorch tensor
transforms.Normalize(mean=[0.21869252622127533], std=[0.1809280514717102]) # Normalize single channel
])
# Apply the transformations and add a batch dimension
image_tensor = transform(image).unsqueeze(0) # Shape: [1, 1, 112, 112]
# Set the device to GPU if available, else CPU
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
image_tensor = image_tensor.to(device)
# Load the NuSPIRe model
model = ViTModel.from_pretrained("TongjiZhanglab/NuSPIRe")
model.to(device)
model.eval() # Set the model to evaluation mode
# Disable gradient calculation for faster inference
with torch.no_grad():
# Perform forward pass through the model
outputs = model(pixel_values=image_tensor)
# Extract the pooled output representation
representation = outputs.pooler_output
# Move the representation to CPU and convert to NumPy array (if further processing is needed)
representation = representation.cpu().numpy()
# Print the output representation
print(representation)
```
### Fine-tuning
NuSPIRe can be fine-tuned on smaller labeled datasets for specific tasks:
```python
# Import necessary libraries
import torch
import requests
from PIL import Image
import lightning.pytorch as pl
from torchvision import transforms
from transformers import ViTForImageClassification
# Set random seed for reproducibility
pl.seed_everything(0, workers=True)
# Open an example image
url = 'https://huggingface.co/TongjiZhanglab/NuSPIRe/resolve/main/Images/image_aabhacci-1.png'
image = Image.open(requests.get(url, stream=True).raw).convert('L')
# Define the image preprocessing transformations
transform = transforms.Compose([
transforms.Resize((112, 112)), # Resize image to 112x112 pixels
transforms.ToTensor(), # Convert PIL image to PyTorch tensor
transforms.Normalize(mean=[0.21869252622127533], std=[0.1809280514717102]) # Normalize single channel
])
# Apply the transformations and add a batch dimension
image_tensor = transform(image).unsqueeze(0) # Shape: [1, 1, 112, 112]
# Set the device to GPU if available, else CPU
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
image_tensor = image_tensor.to(device)
# Load the NuSPIRe model for image classification
model = ViTForImageClassification.from_pretrained("TongjiZhanglab/NuSPIRe", num_labels=2)
model.to(device)
model.train() # Set the model to training mode
# Prepare the labels tensor (example with label 0)
labels = torch.tensor([0]).to(device)
# Forward pass: Compute outputs and loss
outputs = model(image_tensor, labels=labels)
logits = outputs.logits
loss = outputs.loss
# Backward pass: Compute gradients
loss.backward()
# Print the outputs for verification
print("Logits:", logits)
print("Loss:", loss.item())
```
## Citation
If you use NuSPIRe in your research, please cite the following paper:
Hua, Y., Li, S., & Zhang, Y. (2024). NuSPIRe: Nuclear Morphology focused Self-supervised Pretrained model for Image Representations. |