File size: 11,304 Bytes
0a28c37 d527680 0a28c37 d527680 7509edd 336fa6d 42701b7 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 7509edd d527680 278d188 d527680 8891e36 d527680 278d188 d527680 7509edd d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 42701b7 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 fb72a96 d527680 4460856 d527680 4460856 d527680 4460856 d527680 4460856 d527680 fb72a96 d527680 |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 |
---
license: mit
tags:
- biology
- fish
- images
- traits
pipeline_tag: image-segmentation
---
# Model Card for Model ID
This model takes in an image of a fish and segments out traits, as described [below](#Trait-List)
## Model Details
`Trained_model_SM.pth` is the fish segmentation model.
`se_resnext50_32x4d-a260b3a4.pth` is a pretrained ConvNets for pytorch ResNeXt used by BGNN-trait-segmentation.
See [github.com/Cadene/pretrained-models.pytorch#resnext](https://github.com/Cadene/pretrained-models.pytorch#resnext) for documentation about the source.
The segmentation model was first trained on ImageNet ([Deng et al., 2009](https://doi.org/10.1109/CVPR.2009.5206848)), and then the model was fine-tuned on a specific set of image data relevant to the domain: [Illinois Natural History Survey Fish Collection](https://fish.inhs.illinois.edu/) (INHS Fish).
The Feature Pyramid Network (FPN) architecture was used for fine-tuning, since it is a CNN-based architecture designed to handle multi-scale feature maps (Lin et al., 2017: [IEEE](https://doi.org/10.1109/CVPR.2017.106), [arXiv](https://doi.org/10.48550/arXiv.1612.03144)).
The FPN uses SE-ResNeXt as the base network (Hu et al., 2018: [IEEE](https://doi.org/10.1109/CVPR.2018.00745), [arXiv](https://doi.org/10.48550/arXiv.1709.01507)).
### Model Description
PyTorch implementation of Fish trait segmentation model.
This segmentation model is based on pretrained model using the [segementation models torch](https://github.com/qubvel/segmentation_models.pytorch).
Then the model is fine tuned on fish images in order to identify (segment) the different traits.
#### Trait list:
```
background
dorsal_fin
adipos_fin
caudal_fin
anal_fin
pelvic_fin
pectoral_fin
head
eye
caudal_fin_ray
alt_fin_ray
trunk
```
- **Developed by:** M. Maruf and Anuj Karpatne
<!--- **Shared by [optional]:** [More Information Needed]-->
<!--- **Model type:** [More Information Needed]-->
<!--- **Language(s) (NLP):** [More Information Needed]-->
- **License:** MIT
<!--- **Finetuned from model [optional]:** [More Information Needed]-->
### Model Sources
<!-- Provide the basic links for the model. -->
- **Repository:** [BGNN-trait-segmentation](https://github.com/hdr-bgnn/BGNN-trait-segmentation)
- **Paper:** [In progress]
<!--- **Demo [optional]:** [More Information Needed]-->
<!-- ## Uses
<!-- Address questions around how the model is intended to be used, including the foreseeable users of the model and those affected by the model. -->
<!-- ### Direct Use
<!-- This section is for the model use without fine-tuning or plugging into a larger ecosystem/app. -->
<!-- [More Information Needed]
<!-- ### Downstream Use [optional]
<!-- This section is for the model use when fine-tuned for a task, or when plugged into a larger ecosystem/app -->
<!-- [More Information Needed]
<!-- ### Out-of-Scope Use
<!-- This section addresses misuse, malicious use, and uses that the model will not work well for. -->
<!-- [More Information Needed]
<!-- ## Bias, Risks, and Limitations
<!-- This section is meant to convey both technical and sociotechnical limitations. -->
<!-- [More Information Needed] -->
<!-- ### Recommendations -->
<!-- This section is meant to convey recommendations with respect to the bias, risk, and technical limitations. -->
<!-- Users (both direct and downstream) should be made aware of the risks, biases and limitations of the model. More information needed for further recommendations. -->
## How to Get Started with the Model
<!--Use the code below to get started with the model.-->
See instructions for use [here](https://github.com/hdr-bgnn/BGNN-trait-segmentation/blob/main/Segment_mini/README.md).
## Training Details
The image data were annotated using [SlicerMorph](https://slicermorph.github.io/) ([Rolfe et al., 2021](https://doi.org/10.1111/2041-210X.13669)) by collaborators W. Dahdul and K. Diamond.
### Training Data
<!-- This should link to a Data Card, perhaps with a short stub of information on what the training data is all about as well as documentation related to data pre-processing or additional filtering. -->
<!--We only have 295 training images, 99 testing images, and 98 validation images. -->
To increase the size and diversity of the training dataset (originally 295 images), we employed data augmentation techniques such as flipping, shifting, rotating, scaling, and adding noise to the original image data to increase the dataset 10-fold.
We developed 12 target classes, or trait masks, for our segmentation problem, each representing different morphological traits of a fish specimen.
The segmentation classes are: dorsal fin, adipose fin, caudal fin, anal fin, pelvic fin, pectoral fin, head minus the eye, eye, caudal fin-ray, alt fin-ray, alt fin-spine, and trunk.
Although minnows do not have adipose fins, the segmentation model was trained on a variety of fish image data, some of which had adipose fins.
We retained this class because the segmentation model may erroneously assign an adipose fin to a minnow (Fig. S1), and a domain scientist examining these outputs may want to analyze the accuracy of the model.
|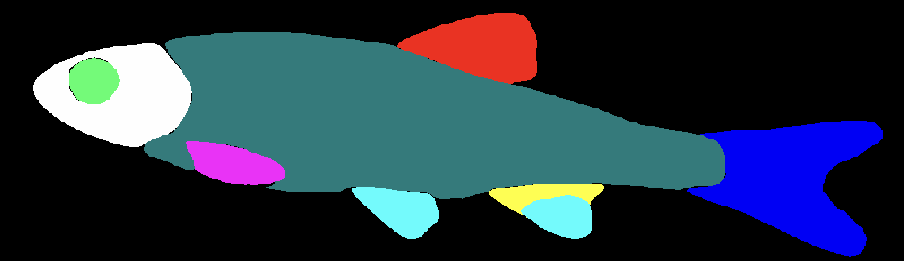|
|:--|
|**Figure S1.** [Image of a segmented minnow](https://huggingface.co/imageomics/BGNN-trait-segmentation/raw/main/Fig_S1.png) with an erroneously marked anal fin.
The segmentation has 13 classes: background (black), dorsal fin (red), adipose fin (green), caudal fin (blue), anal fin (yellow), pelvic fin (light blue), pectoral fin (pink), head minus the eye (white), eye (bright green), trunk (teal), caudal fin-ray (light red), alt fin-ray (light pink), and alt fin-spine (peach).|
The training dataset utilized the image files listed in [training_dataset_INHS.txt](https://huggingface.co/imageomics/BGNN-trait-segmentation/blob/main/training_dataset_INHS.txt).
The validation dataset utilized the image files listed in [validation_dataset_INHS.txt](https://huggingface.co/imageomics/BGNN-trait-segmentation/blob/main/validation_dataset_INHS.txt).
### Training Procedure
<!-- This relates heavily to the Technical Specifications. Content here should link to that section when it is relevant to the training procedure. -->
We prepared the model by using the [Segmentation Model PyTorch library](https://segmentation-modelspytorch.readthedocs.io/en/latest/) (Iakubovskii, 2019) to load an FPN segmentation model that was pretrained on the Imagenet dataset.
We used SE-ResNeXt as the base network/encoder to extract features (embedding) from the input image data and replaced the last decoder layer with 12 target classes.
During the fine-tuning procedure, the encoder of the pre-trained model was frozen as these layers already contain useful features that we can leverage.
We only tuned the decoder weights of our segmentation model during this fine-tuning procedure.
We then trained the prepared model for 120 epochs, updating the weights using dice loss as a measure of similarity between the predicted and ground-truth segmentation.
The Adam optimizer ([Kingma & Ba, 2014](https://doi.org/10.48550/arXiv.1412.6980)) with a small learning rate (1e-4) was used to update the model weights.
<!-- #### Preprocessing [optional]
<!-- [More Information Needed]
<!-- #### Training Hyperparameters
<!-- - **Training regime:** [More Information Needed] <!--fp32, fp16 mixed precision, bf16 mixed precision, bf16 non-mixed precision, fp16 non-mixed precision, fp8 mixed precision -->
<!-- #### Speeds, Sizes, Times [optional]
<!-- This section provides information about throughput, start/end time, checkpoint size if relevant, etc. -->
<!-- [More Information Needed] -->
## Evaluation
<!-- This section describes the evaluation protocols and provides the results. -->
We evaluated the performance of the fine-tuned segmentation model on the test set using the Intersection over Union (IoU) score with a 0.5 threshold.
(The IoU score ranges from 0 to 1, with 1 indicating a perfect overlap between the predicted segmentation and the ground-truth segmentation and 0 indicating no overlap.)
Our segmentation model achieved a 0.90 mIoU score on the test dataset.
### Testing Data, Factors & Metrics
We had 99 testing images and 98 validation images.
<!-- #### Testing Data
<!-- This should link to a Data Card if possible. -->
<!-- [More Information Needed]
<!-- #### Factors
<!-- These are the things the evaluation is disaggregating by, e.g., subpopulations or domains. -->
<!-- [More Information Needed]
<!-- #### Metrics
<!-- These are the evaluation metrics being used, ideally with a description of why. -->
<!-- [More Information Needed]
<!-- ### Results
<!-- [More Information Needed]
<!-- #### Summary
<!-- ## Model Examination [optional]
<!-- Relevant interpretability work for the model goes here -->
<!-- [More Information Needed]
<!-- ## Environmental Impact
<!-- Total emissions (in grams of CO2eq) and additional considerations, such as electricity usage, go here. Edit the suggested text below accordingly -->
<!-- Carbon emissions can be estimated using the [Machine Learning Impact calculator](https://mlco2.github.io/impact#compute) presented in [Lacoste et al. (2019)](https://doi.org/10.48550/arXiv.1910.09700).
- **Hardware Type:** [More Information Needed]
- **Hours used:** [More Information Needed]
- **Cloud Provider:** [More Information Needed]
- **Compute Region:** [More Information Needed]
- **Carbon Emitted:** [More Information Needed] -->
<!-- ## Technical Specifications [optional]
<!-- ### Model Architecture and Objective
<!-- [More Information Needed]
<!-- ### Compute Infrastructure
<!-- [More Information Needed]
<!-- #### Hardware
<!-- [More Information Needed]
<!-- #### Software
<!-- [More Information Needed] -->
<!-- ## Citation [optional]
<!-- If there is a paper or blog post introducing the model, the APA and Bibtex information for that should go in this section. -->
<!-- M. Maruf, 2023, "BGNN-trait-segmentation", DOI coming with model soon for paper citation -->
<!-- **BibTeX:**
[More Information Needed]
**APA:**
[More Information Needed] -->
<!-- ## Glossary [optional]
<!-- If relevant, include terms and calculations in this section that can help readers understand the model or model card. -->
<!-- [More Information Needed] -->
## More Information
Research supported by NSF Office of Advanced Cyberinfrastructure (OAC) Awards [#2022042](https://www.nsf.gov/awardsearch/showAward?AWD_ID=2022042&HistoricalAwards=false), [#1940233](https://nsf.gov/awardsearch/showAward?AWD_ID=1940233&HistoricalAwards=false), [#1940322](https://www.nsf.gov/awardsearch/showAward?AWD_ID=1940322&HistoricalAwards=false), and [#1940247](https://www.nsf.gov/awardsearch/showAward?AWD_ID=1940247&HistoricalAwards=false), with additional support from NSF [Award #2118240](https://www.nsf.gov/awardsearch/showAward?AWD_ID=2118240).
Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
<!-- ## Model Card Authors [optional]
[More Information Needed]
## Model Card Contact
[More Information Needed] -->
|