update benchmark on A100
Browse files- README.md +13 -13
- configs/metadata.json +4 -3
- configs/multi_gpu_evaluate.json +1 -1
- configs/multi_gpu_train.json +1 -1
- docs/README.md +13 -13
- models/model.pt +1 -1
- models/model.ts +2 -2
README.md
CHANGED
|
@@ -21,7 +21,7 @@ The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovern
|
|
| 21 |
wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
|
| 22 |
unzip -q consep_dataset.zip
|
| 23 |
```
|
| 24 |
-
:
|
| 116 |
|
| 117 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 118 |
|-----------|--------|--------------|------------|----------------|
|
| 119 |
-
| Precision | 0.
|
| 120 |
-
| Recall | 0.
|
| 121 |
-
| F1-score | 0.
|
| 122 |
|
| 123 |
|
| 124 |
<hr/>
|
|
@@ -126,22 +126,22 @@ Confusion Metrics for <b>Training</b> for individual classes are (at epoch 50):
|
|
| 126 |
|
| 127 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 128 |
|-----------|--------|--------------|------------|----------------|
|
| 129 |
-
| Precision | 0.
|
| 130 |
-
| Recall | 0.
|
| 131 |
-
| F1-score | 0.
|
| 132 |
|
| 133 |
|
| 134 |
|
| 135 |
## Training Performance
|
| 136 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 137 |
|
| 138 |
-
<br/>
|
| 25 |
|
| 26 |
## Training configuration
|
| 27 |
The training was performed with the following:
|
|
|
|
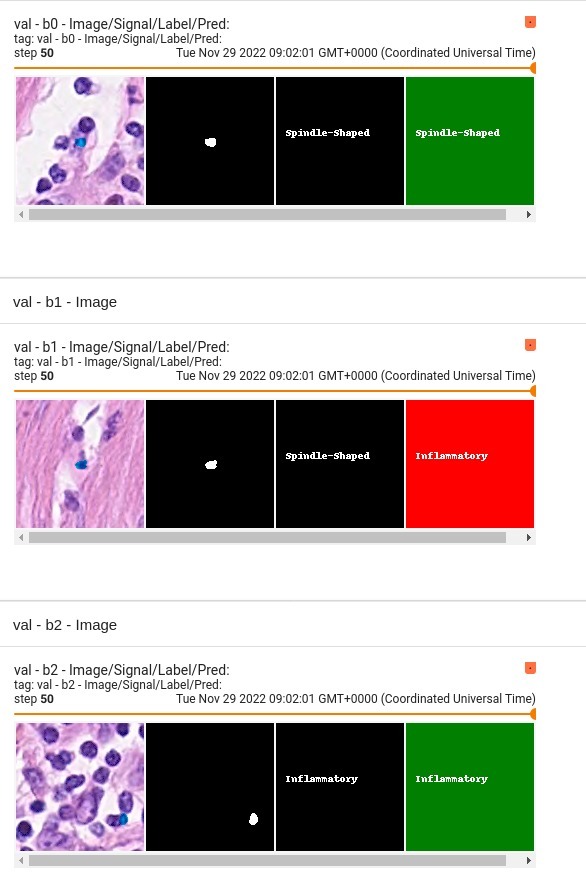
| 103 |
- 2 = Epithelial
|
| 104 |
- 3 = Spindle-Shaped
|
| 105 |
|
| 106 |
+
|
| 107 |
|
| 108 |
## Scores
|
| 109 |
This model achieves the following F1 score on the validation data provided as part of the dataset:
|
| 110 |
|
| 111 |
+
- Train F1 score = 0.941
|
| 112 |
+
- Validation F1 score = 0.840
|
| 113 |
|
| 114 |
<hr/>
|
| 115 |
Confusion Metrics for <b>Validation</b> for individual classes are (at epoch 50):
|
| 116 |
|
| 117 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 118 |
|-----------|--------|--------------|------------|----------------|
|
| 119 |
+
| Precision | 0.6250 | 0.7085 | 0.9188 | 0.8571 |
|
| 120 |
+
| Recall | 0.1449 | 0.8750 | 0.9310 | 0.8154 |
|
| 121 |
+
| F1-score | 0.2353 | 0.7830 | 0.9249 | 0.8357 |
|
| 122 |
|
| 123 |
|
| 124 |
<hr/>
|
|
|
|
| 126 |
|
| 127 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 128 |
|-----------|--------|--------------|------------|----------------|
|
| 129 |
+
| Precision | 0.8902 | 0.9418 | 0.9717 | 0.9189 |
|
| 130 |
+
| Recall | 0.7935 | 0.9250 | 0.9725 | 0.9345 |
|
| 131 |
+
| F1-score | 0.8391 | 0.9333 | 0.9721 | 0.9267 |
|
| 132 |
|
| 133 |
|
| 134 |
|
| 135 |
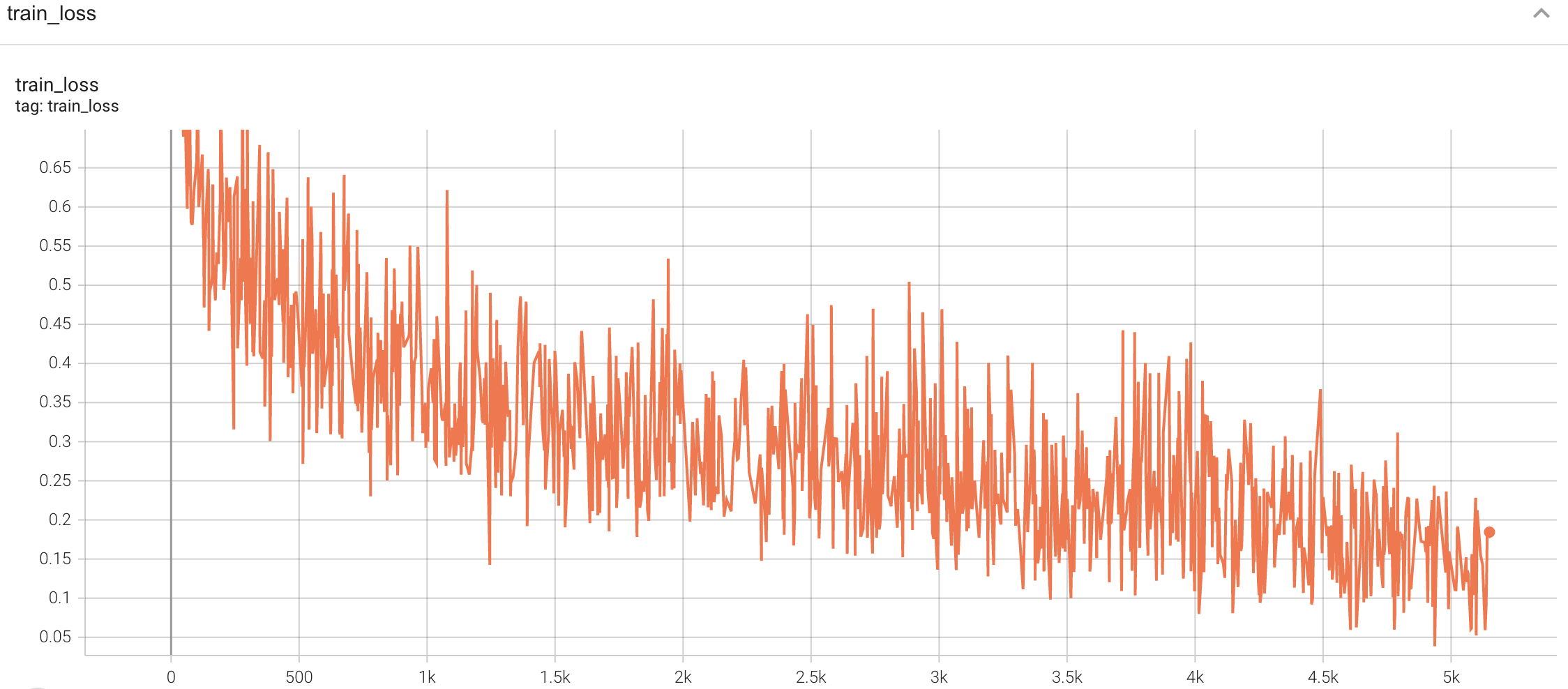
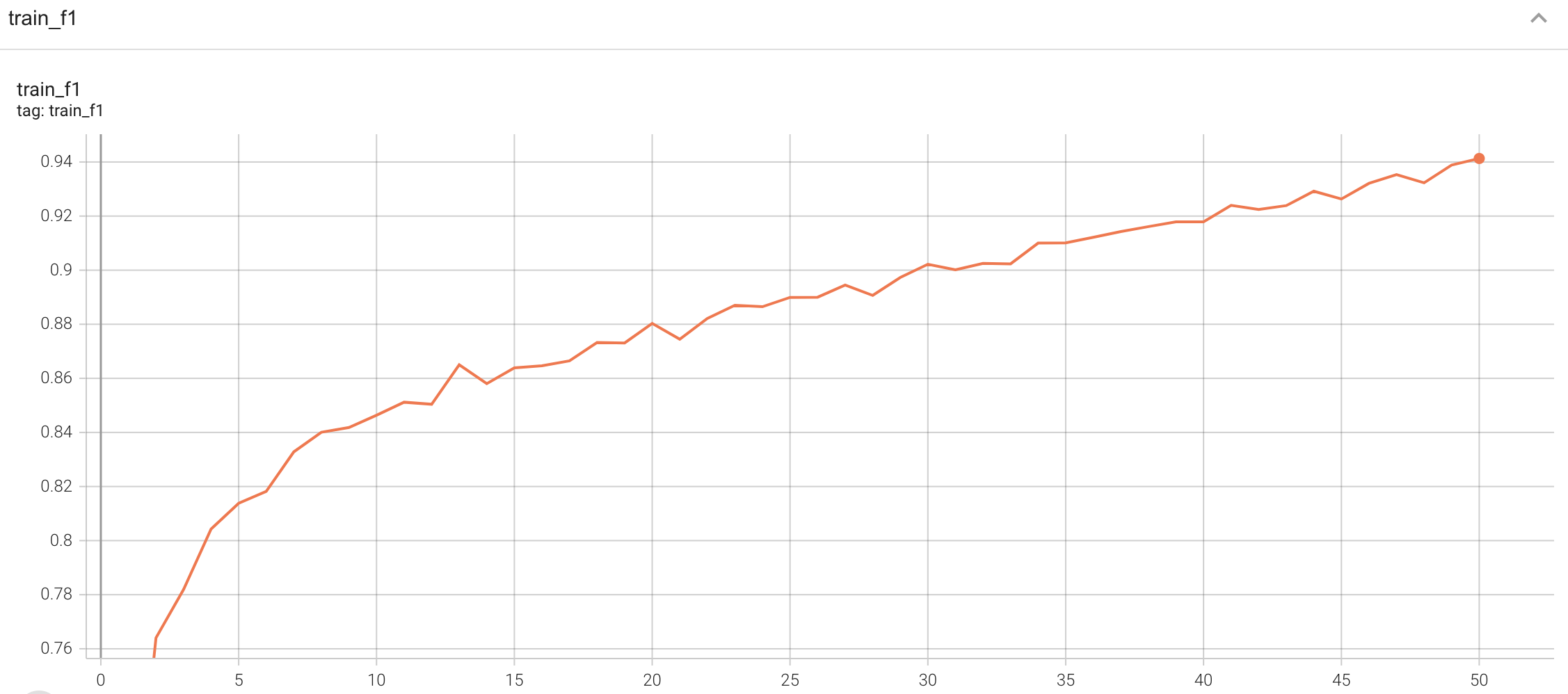
## Training Performance
|
| 136 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 137 |
|
| 138 |
+
 <br>
|
| 139 |
+
 <br>
|
| 140 |
|
| 141 |
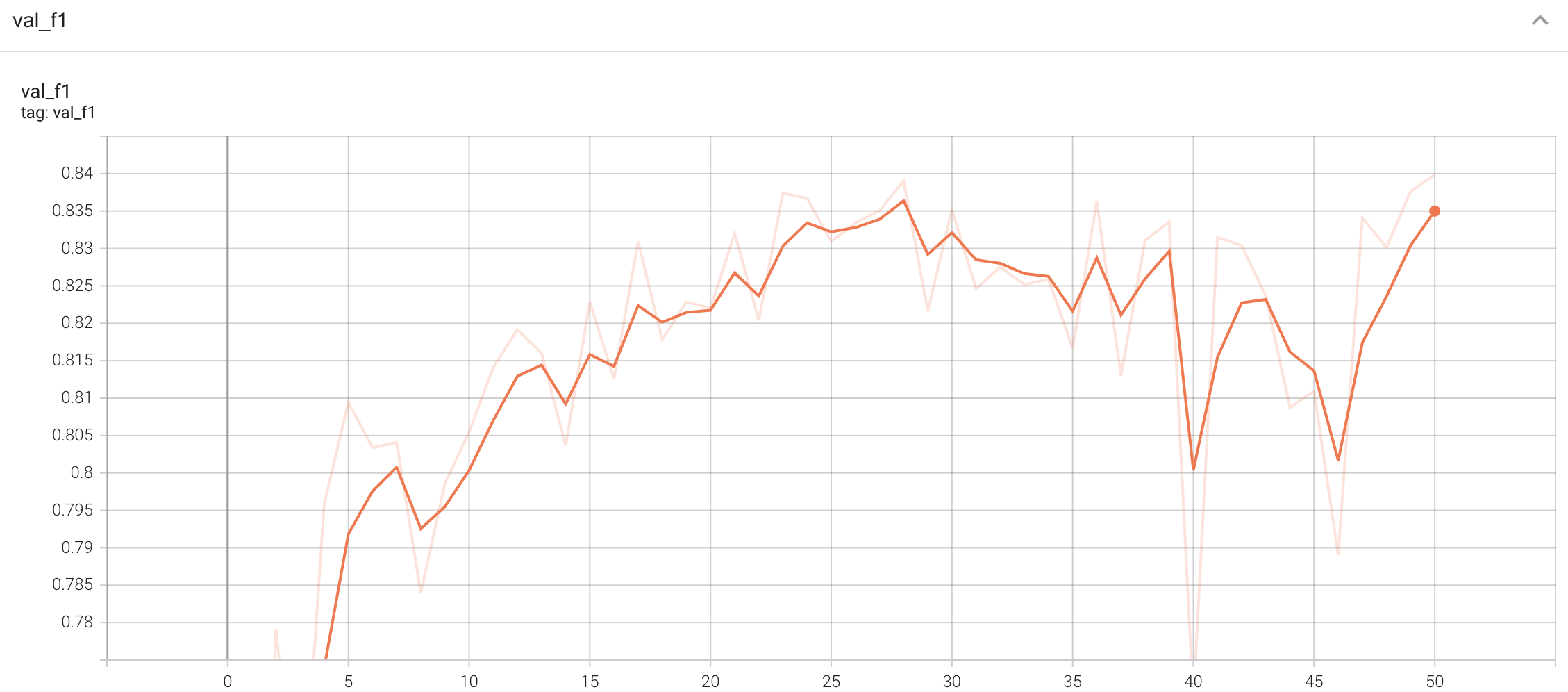
## Validation Performance
|
| 142 |
A graph showing the validation F1-score over 50 epochs.
|
| 143 |
|
| 144 |
+
 <br>
|
| 145 |
|
| 146 |
|
| 147 |
## commands example
|
configs/metadata.json
CHANGED
|
@@ -1,7 +1,8 @@
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
-
"version": "0.0.
|
| 4 |
"changelog": {
|
|
|
|
| 5 |
"0.0.6": "adapt to BundleWorkflow interface",
|
| 6 |
"0.0.5": "add name tag",
|
| 7 |
"0.0.4": "Fix evaluation",
|
|
@@ -9,7 +10,7 @@
|
|
| 9 |
"0.0.2": "Update The Torch Vision Transform",
|
| 10 |
"0.0.1": "initialize the model package structure"
|
| 11 |
},
|
| 12 |
-
"monai_version": "1.2.
|
| 13 |
"pytorch_version": "1.13.1",
|
| 14 |
"numpy_version": "1.22.2",
|
| 15 |
"optional_packages_version": {
|
|
@@ -28,7 +29,7 @@
|
|
| 28 |
"label_classes": "single channel data",
|
| 29 |
"pred_classes": "4 channels OneHot data, channel 0 is Other, channel 1 is Inflammatory, channel 2 is Epithelial, channel 3 is Spindle-Shaped",
|
| 30 |
"eval_metrics": {
|
| 31 |
-
"f1_score": 0.
|
| 32 |
},
|
| 33 |
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 34 |
"references": [
|
|
|
|
| 1 |
{
|
| 2 |
"schema": "https://github.com/Project-MONAI/MONAI-extra-test-data/releases/download/0.8.1/meta_schema_20220324.json",
|
| 3 |
+
"version": "0.0.7",
|
| 4 |
"changelog": {
|
| 5 |
+
"0.0.7": "update benchmark on A100",
|
| 6 |
"0.0.6": "adapt to BundleWorkflow interface",
|
| 7 |
"0.0.5": "add name tag",
|
| 8 |
"0.0.4": "Fix evaluation",
|
|
|
|
| 10 |
"0.0.2": "Update The Torch Vision Transform",
|
| 11 |
"0.0.1": "initialize the model package structure"
|
| 12 |
},
|
| 13 |
+
"monai_version": "1.2.0rc4",
|
| 14 |
"pytorch_version": "1.13.1",
|
| 15 |
"numpy_version": "1.22.2",
|
| 16 |
"optional_packages_version": {
|
|
|
|
| 29 |
"label_classes": "single channel data",
|
| 30 |
"pred_classes": "4 channels OneHot data, channel 0 is Other, channel 1 is Inflammatory, channel 2 is Epithelial, channel 3 is Spindle-Shaped",
|
| 31 |
"eval_metrics": {
|
| 32 |
+
"f1_score": 0.84
|
| 33 |
},
|
| 34 |
"intended_use": "This is an example, not to be used for diagnostic purposes",
|
| 35 |
"references": [
|
configs/multi_gpu_evaluate.json
CHANGED
|
@@ -31,6 +31,6 @@
|
|
| 31 |
"$@validate#evaluator.run()"
|
| 32 |
],
|
| 33 |
"finalize": [
|
| 34 |
-
"$dist.destroy_process_group()"
|
| 35 |
]
|
| 36 |
}
|
|
|
|
| 31 |
"$@validate#evaluator.run()"
|
| 32 |
],
|
| 33 |
"finalize": [
|
| 34 |
+
"$dist.is_initialized() and dist.destroy_process_group()"
|
| 35 |
]
|
| 36 |
}
|
configs/multi_gpu_train.json
CHANGED
|
@@ -40,6 +40,6 @@
|
|
| 40 |
"$@train#trainer.run()"
|
| 41 |
],
|
| 42 |
"finalize": [
|
| 43 |
-
"$dist.destroy_process_group()"
|
| 44 |
]
|
| 45 |
}
|
|
|
|
| 40 |
"$@train#trainer.run()"
|
| 41 |
],
|
| 42 |
"finalize": [
|
| 43 |
+
"$dist.is_initialized() and dist.destroy_process_group()"
|
| 44 |
]
|
| 45 |
}
|
docs/README.md
CHANGED
|
@@ -14,7 +14,7 @@ The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovern
|
|
| 14 |
wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip
|
| 15 |
unzip -q consep_dataset.zip
|
| 16 |
```
|
| 17 |
-
:
|
| 109 |
|
| 110 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 111 |
|-----------|--------|--------------|------------|----------------|
|
| 112 |
-
| Precision | 0.
|
| 113 |
-
| Recall | 0.
|
| 114 |
-
| F1-score | 0.
|
| 115 |
|
| 116 |
|
| 117 |
<hr/>
|
|
@@ -119,22 +119,22 @@ Confusion Metrics for <b>Training</b> for individual classes are (at epoch 50):
|
|
| 119 |
|
| 120 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 121 |
|-----------|--------|--------------|------------|----------------|
|
| 122 |
-
| Precision | 0.
|
| 123 |
-
| Recall | 0.
|
| 124 |
-
| F1-score | 0.
|
| 125 |
|
| 126 |
|
| 127 |
|
| 128 |
## Training Performance
|
| 129 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 130 |
|
| 131 |
-
<br/>
|
| 18 |
|
| 19 |
## Training configuration
|
| 20 |
The training was performed with the following:
|
|
|
|
| 96 |
- 2 = Epithelial
|
| 97 |
- 3 = Spindle-Shaped
|
| 98 |
|
| 99 |
+

|
| 100 |
|
| 101 |
## Scores
|
| 102 |
This model achieves the following F1 score on the validation data provided as part of the dataset:
|
| 103 |
|
| 104 |
+
- Train F1 score = 0.941
|
| 105 |
+
- Validation F1 score = 0.840
|
| 106 |
|
| 107 |
<hr/>
|
| 108 |
Confusion Metrics for <b>Validation</b> for individual classes are (at epoch 50):
|
| 109 |
|
| 110 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 111 |
|-----------|--------|--------------|------------|----------------|
|
| 112 |
+
| Precision | 0.6250 | 0.7085 | 0.9188 | 0.8571 |
|
| 113 |
+
| Recall | 0.1449 | 0.8750 | 0.9310 | 0.8154 |
|
| 114 |
+
| F1-score | 0.2353 | 0.7830 | 0.9249 | 0.8357 |
|
| 115 |
|
| 116 |
|
| 117 |
<hr/>
|
|
|
|
| 119 |
|
| 120 |
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped |
|
| 121 |
|-----------|--------|--------------|------------|----------------|
|
| 122 |
+
| Precision | 0.8902 | 0.9418 | 0.9717 | 0.9189 |
|
| 123 |
+
| Recall | 0.7935 | 0.9250 | 0.9725 | 0.9345 |
|
| 124 |
+
| F1-score | 0.8391 | 0.9333 | 0.9721 | 0.9267 |
|
| 125 |
|
| 126 |
|
| 127 |
|
| 128 |
## Training Performance
|
| 129 |
A graph showing the training Loss and F1-score over 50 epochs.
|
| 130 |
|
| 131 |
+
 <br>
|
| 132 |
+
 <br>
|
| 133 |
|
| 134 |
## Validation Performance
|
| 135 |
A graph showing the validation F1-score over 50 epochs.
|
| 136 |
|
| 137 |
+
 <br>
|
| 138 |
|
| 139 |
|
| 140 |
## commands example
|
models/model.pt
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
size 28419489
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:f9aef3d7c3114f1c86695e119d64f7e83dcf6449716cb64df9d5b7b447adb8a5
|
| 3 |
size 28419489
|
models/model.ts
CHANGED
|
@@ -1,3 +1,3 @@
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
-
oid sha256:
|
| 3 |
-
size
|
|
|
|
| 1 |
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:37d348addc879bc51d5e5fa89927ceb48c3521632135a0d5cdc488592961209e
|
| 3 |
+
size 28620879
|