deploy at 2024-12-02 13:24:21.737970
Browse files- .gitignore +3 -0
- Dockerfile +10 -0
- README.md +6 -5
- acr24.py +0 -0
- config.ini +6 -0
- main.py +461 -0
- media/favicon.ico +0 -0
- media/logo_large.png +0 -0
- media/me.jpeg +0 -0
- posts/2022-01-25-unbuilt-drug-safety-australias-digital-health-infrastructure.md +34 -0
- posts/2022-01-29-machine-learning-to-get-stuff-done.md +81 -0
- posts/images/BPE.drawio.svg +4 -0
- posts/images/Recurrent_neural_network_unfold.svg +793 -0
- posts/images/paste-35A12A44.png +0 -0
- posts/images/prodigy.svg +0 -0
- posts/images/unnamed-chunk-1-1.png +0 -0
- posts/images/unnamed-chunk-11-1.png +0 -0
- posts/images/unnamed-chunk-15-1.png +0 -0
- posts/images/unnamed-chunk-16-1.png +0 -0
- posts/images/unnamed-chunk-7-1.png +0 -0
- posts/images/unnamed-chunk-9-1.png +0 -0
- requirements.txt +4 -0
- static/css/style.css +195 -0
.gitignore
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
/.DS_Store
|
| 2 |
+
/__pycache__
|
| 3 |
+
.sesskey
|
Dockerfile
ADDED
|
@@ -0,0 +1,10 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
FROM python:3.10
|
| 2 |
+
WORKDIR /code
|
| 3 |
+
COPY --link --chown=1000 . .
|
| 4 |
+
RUN mkdir -p /tmp/cache/
|
| 5 |
+
RUN chmod a+rwx -R /tmp/cache/
|
| 6 |
+
ENV HF_HUB_CACHE=HF_HOME
|
| 7 |
+
RUN pip install --no-cache-dir -r requirements.txt
|
| 8 |
+
|
| 9 |
+
ENV PYTHONUNBUFFERED=1 PORT=7860
|
| 10 |
+
CMD ["python", "main.py"]
|
README.md
CHANGED
|
@@ -1,10 +1,11 @@
|
|
|
|
|
| 1 |
---
|
| 2 |
-
title:
|
| 3 |
-
emoji:
|
| 4 |
-
colorFrom:
|
| 5 |
colorTo: red
|
| 6 |
sdk: docker
|
|
|
|
| 7 |
pinned: false
|
|
|
|
| 8 |
---
|
| 9 |
-
|
| 10 |
-
Check out the configuration reference at https://huggingface.co/docs/hub/spaces-config-reference
|
|
|
|
| 1 |
+
|
| 2 |
---
|
| 3 |
+
title: cmcmaster/website
|
| 4 |
+
emoji: 🚀
|
| 5 |
+
colorFrom: purple
|
| 6 |
colorTo: red
|
| 7 |
sdk: docker
|
| 8 |
+
app_file: app.py
|
| 9 |
pinned: false
|
| 10 |
+
termination_grace_period: 2m
|
| 11 |
---
|
|
|
|
|
|
acr24.py
ADDED
|
File without changes
|
config.ini
ADDED
|
@@ -0,0 +1,6 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
[DEFAULT]
|
| 2 |
+
dataset_id = space-backup
|
| 3 |
+
db_dir = data
|
| 4 |
+
private_backup = True
|
| 5 |
+
interval = 15
|
| 6 |
+
|
main.py
ADDED
|
@@ -0,0 +1,461 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from fasthtml.common import *
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
from datetime import datetime
|
| 4 |
+
from starlette.responses import RedirectResponse, FileResponse
|
| 5 |
+
from starlette.requests import Request
|
| 6 |
+
import json
|
| 7 |
+
import numpy as np
|
| 8 |
+
from sklearn.metrics.pairwise import cosine_similarity
|
| 9 |
+
from fasthtml_hf import setup_hf_backup
|
| 10 |
+
|
| 11 |
+
# Load projects from JSON file
|
| 12 |
+
with open('data/projects.json', 'r') as f:
|
| 13 |
+
projects = json.load(f)
|
| 14 |
+
|
| 15 |
+
# Load summaries and abstracts
|
| 16 |
+
with open("data/acr/2024/summaries.json") as f:
|
| 17 |
+
summaries = json.loads(f.read())
|
| 18 |
+
|
| 19 |
+
with open("data/acr/2024/abstracts.jsonl", "r") as f:
|
| 20 |
+
abstracts = [json.loads(line) for line in f]
|
| 21 |
+
|
| 22 |
+
for abstract in abstracts:
|
| 23 |
+
abstract['abstract'] = abstract['abstract'].replace("## Background/Purpose\n", "Background/Purpose\n")
|
| 24 |
+
abstract['abstract'] = abstract['abstract'].replace("## Methods\n", "Methods\n")
|
| 25 |
+
|
| 26 |
+
# Load embeddings
|
| 27 |
+
embeddings = np.load('data/acr/2024/embeddings.npy')
|
| 28 |
+
|
| 29 |
+
# Associate embeddings with abstracts
|
| 30 |
+
for abstract, embedding in zip(abstracts, embeddings):
|
| 31 |
+
abstract['embedding'] = embedding
|
| 32 |
+
|
| 33 |
+
# Setup app with headers
|
| 34 |
+
hdrs = (
|
| 35 |
+
picolink,
|
| 36 |
+
StyleX("static/css/style.css"),
|
| 37 |
+
MarkdownJS(),
|
| 38 |
+
HighlightJS(langs=['python', 'javascript', 'html', 'css'])
|
| 39 |
+
)
|
| 40 |
+
app = FastHTML(hdrs=hdrs)
|
| 41 |
+
|
| 42 |
+
def get_posts():
|
| 43 |
+
posts = []
|
| 44 |
+
for path in Path("posts").glob("*.md"):
|
| 45 |
+
with open(path, 'r') as f:
|
| 46 |
+
title = f.readline().strip()
|
| 47 |
+
date = datetime.fromtimestamp(path.stat().st_mtime)
|
| 48 |
+
posts.append({"title": title, "date": date, "path": path})
|
| 49 |
+
return sorted(posts, key=lambda x: x['date'], reverse=True)
|
| 50 |
+
|
| 51 |
+
def nav_menu(current_page="home"):
|
| 52 |
+
return Nav(
|
| 53 |
+
A("Home", href="/", cls="active" if current_page == "home" else ""),
|
| 54 |
+
A("Blog", href="/blog", cls="active" if current_page == "blog" else ""),
|
| 55 |
+
A("Projects", href="/projects", cls="active" if current_page == "projects" else ""),
|
| 56 |
+
A("ACR24", href="/acr24", cls="active" if current_page == "acr24" else ""),
|
| 57 |
+
)
|
| 58 |
+
|
| 59 |
+
# Routes
|
| 60 |
+
@app.get("/")
|
| 61 |
+
def home():
|
| 62 |
+
content = Main(
|
| 63 |
+
nav_menu("home"),
|
| 64 |
+
Div(
|
| 65 |
+
Img(src="media/me.jpeg", alt="Profile Photo"),
|
| 66 |
+
H1("Dr. Chris McMaster"),
|
| 67 |
+
P("Rheumatologist and Data Scientist"),
|
| 68 |
+
P("Using AI to improve healthcare"),
|
| 69 |
+
cls="profile"
|
| 70 |
+
),
|
| 71 |
+
cls="container"
|
| 72 |
+
)
|
| 73 |
+
return Title("RheumAI"), content
|
| 74 |
+
|
| 75 |
+
@app.get("/blog")
|
| 76 |
+
def blog():
|
| 77 |
+
posts = get_posts()
|
| 78 |
+
content = Main(
|
| 79 |
+
nav_menu("blog"),
|
| 80 |
+
Div(
|
| 81 |
+
H1("Blog Posts"),
|
| 82 |
+
*[Div(
|
| 83 |
+
H2(A(post['title'], href=f"/post/{post['path'].stem}")),
|
| 84 |
+
P(post['date'].strftime('%Y-%m-%d'), cls="date"),
|
| 85 |
+
cls="blog-post"
|
| 86 |
+
) for post in posts],
|
| 87 |
+
cls="container"
|
| 88 |
+
)
|
| 89 |
+
)
|
| 90 |
+
return Title("Blog - RheumAI"), content
|
| 91 |
+
|
| 92 |
+
@app.get("/projects")
|
| 93 |
+
def projects_page():
|
| 94 |
+
content = Main(
|
| 95 |
+
nav_menu("projects"),
|
| 96 |
+
Div(
|
| 97 |
+
H1("Projects"),
|
| 98 |
+
*[Div(
|
| 99 |
+
H2(project["name"]),
|
| 100 |
+
P(project["description"]),
|
| 101 |
+
A("Visit Project", href=project["external_link"], cls="button"),
|
| 102 |
+
cls="blog-post"
|
| 103 |
+
) for project in projects],
|
| 104 |
+
cls="container"
|
| 105 |
+
)
|
| 106 |
+
)
|
| 107 |
+
return Title("Projects - RheumAI"), content
|
| 108 |
+
|
| 109 |
+
# Static file handlers
|
| 110 |
+
app.mount("/static", StaticFiles(directory="static"), name="static")
|
| 111 |
+
app.mount("/media", StaticFiles(directory="media"), name="media")
|
| 112 |
+
app.mount("/posts/images", StaticFiles(directory="posts/images"), name="post_images")
|
| 113 |
+
app.mount("/data/acr/2024", StaticFiles(directory="data/acr/2024"), name="acr24_data")
|
| 114 |
+
|
| 115 |
+
@app.get("/post/{slug}")
|
| 116 |
+
def post(slug: str):
|
| 117 |
+
path = Path(f"posts/{slug}.md")
|
| 118 |
+
if not path.exists():
|
| 119 |
+
return "Post not found", 404
|
| 120 |
+
|
| 121 |
+
with open(path, 'r') as f:
|
| 122 |
+
title = f.readline().strip()
|
| 123 |
+
content = f.read()
|
| 124 |
+
|
| 125 |
+
# Fix image paths - replace both relative and absolute paths
|
| 126 |
+
content = content.replace('](images/', '](/posts/images/')
|
| 127 |
+
content = content.replace('](/images/', '](/posts/images/')
|
| 128 |
+
|
| 129 |
+
# Add styling for blog post images
|
| 130 |
+
img_style = Style("""
|
| 131 |
+
.blog-post img {
|
| 132 |
+
max-width: 100%;
|
| 133 |
+
height: auto;
|
| 134 |
+
margin: 1em 0;
|
| 135 |
+
}
|
| 136 |
+
.blog-post .marked {
|
| 137 |
+
overflow-x: auto;
|
| 138 |
+
}
|
| 139 |
+
""")
|
| 140 |
+
|
| 141 |
+
# Find the current abstract
|
| 142 |
+
current_abstract = next((a for a in abstracts if a['slug'] == slug), None)
|
| 143 |
+
if not current_abstract:
|
| 144 |
+
return "Abstract not found", 404
|
| 145 |
+
|
| 146 |
+
# Add a button to find similar abstracts
|
| 147 |
+
similar_button = Button("Find Similar Abstracts",
|
| 148 |
+
hx_get=f"/acr24/similar/{slug}",
|
| 149 |
+
hx_target="#similar-abstracts",
|
| 150 |
+
cls="button")
|
| 151 |
+
|
| 152 |
+
content = content.replace('</div>', f'\n<div id="similar-abstracts"></div>\n{similar_button}</div>')
|
| 153 |
+
|
| 154 |
+
return Title(title), (
|
| 155 |
+
img_style,
|
| 156 |
+
Main(
|
| 157 |
+
nav_menu("blog"),
|
| 158 |
+
Div(
|
| 159 |
+
H1(title),
|
| 160 |
+
Div(content, cls="marked"),
|
| 161 |
+
cls="blog-post container"
|
| 162 |
+
)
|
| 163 |
+
)
|
| 164 |
+
)
|
| 165 |
+
|
| 166 |
+
@app.get("/favicon.ico")
|
| 167 |
+
def favicon():
|
| 168 |
+
return FileResponse("static/favicon.ico")
|
| 169 |
+
|
| 170 |
+
|
| 171 |
+
|
| 172 |
+
|
| 173 |
+
@app.get("/acr24")
|
| 174 |
+
def acr24():
|
| 175 |
+
# Load the summaries from JSON file
|
| 176 |
+
with open("data/acr/2024/summaries.json") as f:
|
| 177 |
+
summaries = json.loads(f.read())
|
| 178 |
+
|
| 179 |
+
|
| 180 |
+
# Create the title and header
|
| 181 |
+
title = Title("ACR 2024")
|
| 182 |
+
header = H1("ACR 2024")
|
| 183 |
+
|
| 184 |
+
# Create tabs with data-tab attributes
|
| 185 |
+
tabs = Nav(
|
| 186 |
+
Ul(
|
| 187 |
+
Li(A("AI Summaries", href="#", data_tab="summaries", cls="active")),
|
| 188 |
+
Li(A("Search", href="#", data_tab="search")),
|
| 189 |
+
Li(A("Embeddings", href="#", data_tab="embeddings")),
|
| 190 |
+
cls="tabs",
|
| 191 |
+
style="margin-bottom: 0"
|
| 192 |
+
)
|
| 193 |
+
)
|
| 194 |
+
|
| 195 |
+
# Add PDF download link to summaries section
|
| 196 |
+
summaries_section = Section(
|
| 197 |
+
H2("AI-Generated Topic Summaries"),
|
| 198 |
+
A("Download as PDF", href="data/acr/2024/summaries.pdf", cls="button", style="margin-bottom: 1rem"),
|
| 199 |
+
Div(*[
|
| 200 |
+
Article(
|
| 201 |
+
H3(topic),
|
| 202 |
+
Div(summary, cls="marked"),
|
| 203 |
+
cls="summary-card"
|
| 204 |
+
) for topic, summary in summaries.items()
|
| 205 |
+
]),
|
| 206 |
+
id="summaries",
|
| 207 |
+
data_section="summaries",
|
| 208 |
+
cls="active"
|
| 209 |
+
)
|
| 210 |
+
|
| 211 |
+
search_section = Section(
|
| 212 |
+
H2("Search Abstracts"),
|
| 213 |
+
Form(
|
| 214 |
+
Input(
|
| 215 |
+
type="search",
|
| 216 |
+
name="q",
|
| 217 |
+
placeholder="Search abstracts...",
|
| 218 |
+
hx_post="/acr24/search",
|
| 219 |
+
hx_trigger="keyup changed delay:500ms",
|
| 220 |
+
hx_target="#search-results"
|
| 221 |
+
),
|
| 222 |
+
cls="search-form"
|
| 223 |
+
),
|
| 224 |
+
Div(id="search-results"),
|
| 225 |
+
id="search",
|
| 226 |
+
data_section="search"
|
| 227 |
+
)
|
| 228 |
+
|
| 229 |
+
# Add CSS for tab functionality
|
| 230 |
+
style = Style("""
|
| 231 |
+
.tabs {
|
| 232 |
+
margin-bottom: 2rem;
|
| 233 |
+
list-style: none;
|
| 234 |
+
padding: 0;
|
| 235 |
+
}
|
| 236 |
+
.tabs li {
|
| 237 |
+
display: inline-block;
|
| 238 |
+
margin-right: 1rem;
|
| 239 |
+
}
|
| 240 |
+
.tabs a {
|
| 241 |
+
display: inline-block;
|
| 242 |
+
padding: 0.5rem 1rem;
|
| 243 |
+
text-decoration: none;
|
| 244 |
+
border: 1px solid transparent;
|
| 245 |
+
border-bottom: none;
|
| 246 |
+
margin-bottom: -1px;
|
| 247 |
+
}
|
| 248 |
+
|
| 249 |
+
[data-section] {
|
| 250 |
+
display: none;
|
| 251 |
+
}
|
| 252 |
+
[data-section].active {
|
| 253 |
+
display: block;
|
| 254 |
+
}
|
| 255 |
+
""")
|
| 256 |
+
|
| 257 |
+
# Update JavaScript to toggle section visibility using classes
|
| 258 |
+
script = Script("""
|
| 259 |
+
document.addEventListener('DOMContentLoaded', function() {
|
| 260 |
+
const tabLinks = document.querySelectorAll('a[data-tab]');
|
| 261 |
+
const sections = document.querySelectorAll('[data-section]');
|
| 262 |
+
|
| 263 |
+
function switchTab(targetTab) {
|
| 264 |
+
// Update active tab
|
| 265 |
+
tabLinks.forEach(link => {
|
| 266 |
+
link.classList.remove('active');
|
| 267 |
+
if (link.dataset.tab === targetTab) {
|
| 268 |
+
link.classList.add('active');
|
| 269 |
+
}
|
| 270 |
+
});
|
| 271 |
+
|
| 272 |
+
// Show/hide sections using classes
|
| 273 |
+
sections.forEach(section => {
|
| 274 |
+
section.classList.remove('active');
|
| 275 |
+
if (section.dataset.section === targetTab) {
|
| 276 |
+
section.classList.add('active');
|
| 277 |
+
}
|
| 278 |
+
});
|
| 279 |
+
}
|
| 280 |
+
|
| 281 |
+
// Add click handlers to tab links
|
| 282 |
+
tabLinks.forEach(link => {
|
| 283 |
+
link.addEventListener('click', (e) => {
|
| 284 |
+
e.preventDefault();
|
| 285 |
+
switchTab(link.dataset.tab);
|
| 286 |
+
});
|
| 287 |
+
});
|
| 288 |
+
|
| 289 |
+
// Set initial active tab
|
| 290 |
+
switchTab('summaries');
|
| 291 |
+
});
|
| 292 |
+
""")
|
| 293 |
+
|
| 294 |
+
# Add embeddings section
|
| 295 |
+
embeddings_section = Section(
|
| 296 |
+
H2("Embeddings Plot"),
|
| 297 |
+
A(Img(src="/data/acr/2024/embeddings.png", alt="Embeddings TSNE Plot"),
|
| 298 |
+
href="/data/acr/2024/embeddings.png"),
|
| 299 |
+
id="embeddings",
|
| 300 |
+
data_section="embeddings"
|
| 301 |
+
)
|
| 302 |
+
|
| 303 |
+
content = Main(
|
| 304 |
+
nav_menu("acr24"),
|
| 305 |
+
Container(
|
| 306 |
+
title,
|
| 307 |
+
header,
|
| 308 |
+
tabs,
|
| 309 |
+
summaries_section,
|
| 310 |
+
search_section,
|
| 311 |
+
embeddings_section, # Add the new section
|
| 312 |
+
style,
|
| 313 |
+
script
|
| 314 |
+
)
|
| 315 |
+
)
|
| 316 |
+
return Title("ACR 2024 - RheumAI"), content
|
| 317 |
+
|
| 318 |
+
@app.post("/acr24/search")
|
| 319 |
+
def search(request: Request, q: str = Form(...)):
|
| 320 |
+
"""Handle abstract search requests"""
|
| 321 |
+
if not q:
|
| 322 |
+
return Div("Enter search terms above")
|
| 323 |
+
|
| 324 |
+
# Convert search query to lowercase for case-insensitive matching
|
| 325 |
+
q = q.lower()
|
| 326 |
+
|
| 327 |
+
# Search through abstracts and find matches
|
| 328 |
+
matches = []
|
| 329 |
+
for abstract in abstracts:
|
| 330 |
+
# Check if query matches in title or content
|
| 331 |
+
if (q in abstract.get('title', '').lower() or
|
| 332 |
+
q in abstract.get('abstract', '').lower()):
|
| 333 |
+
matches.append(abstract)
|
| 334 |
+
|
| 335 |
+
# If no results found
|
| 336 |
+
if not matches:
|
| 337 |
+
return Div(P("No matching abstracts found"))
|
| 338 |
+
|
| 339 |
+
# Create view toggle buttons
|
| 340 |
+
toggle_buttons = Div(
|
| 341 |
+
Button("Show First 10",
|
| 342 |
+
hx_post=f"/acr24/search?q={q}&limit=10",
|
| 343 |
+
hx_target="#search-results",
|
| 344 |
+
cls="active"),
|
| 345 |
+
Button("Show All",
|
| 346 |
+
hx_post=f"/acr24/search?q={q}&limit=all",
|
| 347 |
+
hx_target="#search-results"),
|
| 348 |
+
cls="view-toggle"
|
| 349 |
+
)
|
| 350 |
+
|
| 351 |
+
# Get limit from query params, default to 10
|
| 352 |
+
limit = request.query_params.get('limit', '10')
|
| 353 |
+
results_to_show = matches if limit == 'all' else matches[:10]
|
| 354 |
+
|
| 355 |
+
# Add modal container if it doesn't exist
|
| 356 |
+
modal_html = Div(
|
| 357 |
+
Div(
|
| 358 |
+
Button("×", cls="modal-close", onclick="closeModal()"),
|
| 359 |
+
Div(id="modal-content"),
|
| 360 |
+
cls="modal"
|
| 361 |
+
),
|
| 362 |
+
id="modal-overlay",
|
| 363 |
+
cls="modal-overlay",
|
| 364 |
+
)
|
| 365 |
+
|
| 366 |
+
# Add JavaScript for modal functionality
|
| 367 |
+
modal_script = Script("""
|
| 368 |
+
function showModal(abstractNumber) {
|
| 369 |
+
fetch(`/acr24/similar/${abstractNumber}`)
|
| 370 |
+
.then(response => response.text())
|
| 371 |
+
.then(html => {
|
| 372 |
+
document.getElementById('modal-content').innerHTML = html;
|
| 373 |
+
document.getElementById('modal-overlay').style.display = 'block';
|
| 374 |
+
});
|
| 375 |
+
}
|
| 376 |
+
|
| 377 |
+
function closeModal() {
|
| 378 |
+
document.getElementById('modal-overlay').style.display = 'none';
|
| 379 |
+
}
|
| 380 |
+
|
| 381 |
+
// Close modal when clicking outside
|
| 382 |
+
document.getElementById('modal-overlay').addEventListener('click', function(e) {
|
| 383 |
+
if (e.target === this) {
|
| 384 |
+
closeModal();
|
| 385 |
+
}
|
| 386 |
+
});
|
| 387 |
+
""")
|
| 388 |
+
|
| 389 |
+
# Return formatted results with Find Similar button for each result
|
| 390 |
+
return Div(
|
| 391 |
+
modal_html,
|
| 392 |
+
modal_script,
|
| 393 |
+
toggle_buttons,
|
| 394 |
+
P(f"Found {len(matches)} matching abstracts:"),
|
| 395 |
+
*[Article(
|
| 396 |
+
H5(A(abstract['title'], href=abstract['link'])),
|
| 397 |
+
P(abstract.get('abstract', '')[:300] + "..."),
|
| 398 |
+
Button("Find Similar",
|
| 399 |
+
onclick=f"showModal('{abstract['abstract_number']}')",
|
| 400 |
+
cls="button similar-button"),
|
| 401 |
+
cls="abstract-result"
|
| 402 |
+
) for abstract in results_to_show]
|
| 403 |
+
)
|
| 404 |
+
|
| 405 |
+
# Add some CSS for the toggle buttons
|
| 406 |
+
style = Style("""
|
| 407 |
+
.view-toggle {
|
| 408 |
+
margin-bottom: 1rem;
|
| 409 |
+
}
|
| 410 |
+
.view-toggle button {
|
| 411 |
+
margin-right: 0.5rem;
|
| 412 |
+
}
|
| 413 |
+
.view-toggle button.active {
|
| 414 |
+
background: #4a5568;
|
| 415 |
+
color: white;
|
| 416 |
+
}
|
| 417 |
+
""")
|
| 418 |
+
|
| 419 |
+
# Routes for specific resources
|
| 420 |
+
@app.get("/{project_name}")
|
| 421 |
+
def project(project_name: str):
|
| 422 |
+
project = next((p for p in projects if p["internal_link"] == project_name), None)
|
| 423 |
+
if project:
|
| 424 |
+
return RedirectResponse(url=project["external_link"])
|
| 425 |
+
else:
|
| 426 |
+
return "Project not found", 404
|
| 427 |
+
|
| 428 |
+
@app.get("/acr24/similar/{abstract_number}")
|
| 429 |
+
def find_similar(abstract_number: str):
|
| 430 |
+
# Find the current abstract
|
| 431 |
+
current_abstract = next((a for a in abstracts if a['abstract_number'] == abstract_number), None)
|
| 432 |
+
if not current_abstract:
|
| 433 |
+
return Div("Abstract not found", cls="error")
|
| 434 |
+
|
| 435 |
+
current_embedding = current_abstract['embedding'].reshape(1, -1)
|
| 436 |
+
|
| 437 |
+
# Compute cosine similarity between the current abstract and all others
|
| 438 |
+
similarities = cosine_similarity(current_embedding, embeddings)[0]
|
| 439 |
+
|
| 440 |
+
# Get top 5 similar abstracts excluding itself
|
| 441 |
+
similar_indices = similarities.argsort()[::-1][1:6]
|
| 442 |
+
similar_abstracts = [abstracts[i] for i in similar_indices]
|
| 443 |
+
|
| 444 |
+
# Create a list of similar abstracts
|
| 445 |
+
return Div(
|
| 446 |
+
H2("Similar Abstracts"),
|
| 447 |
+
Ul(
|
| 448 |
+
*[
|
| 449 |
+
Li(
|
| 450 |
+
H3(A(abstract['title'], href=abstract['link'])),
|
| 451 |
+
P(f"Topic: {abstract['topic']}"),
|
| 452 |
+
Div(abstract.get('abstract', '')[:200] + "...", cls="marked"),
|
| 453 |
+
cls="similar-abstract"
|
| 454 |
+
) for abstract in similar_abstracts
|
| 455 |
+
],
|
| 456 |
+
cls="similar-list"
|
| 457 |
+
)
|
| 458 |
+
)
|
| 459 |
+
setup_hf_backup(app)
|
| 460 |
+
if __name__ == "__main__":
|
| 461 |
+
serve()
|
media/favicon.ico
ADDED
|
|
media/logo_large.png
ADDED

|
media/me.jpeg
ADDED

|
posts/2022-01-25-unbuilt-drug-safety-australias-digital-health-infrastructure.md
ADDED
|
@@ -0,0 +1,34 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Drug Safety & Australia’s Digital Health Infrastructure
|
| 2 |
+
|
| 3 |
+
## Missing Data and Missing the Mark
|
| 4 |
+
|
| 5 |
+
John wakes up one morning with sharp chest pains, which he dismisses as just another nuisance pain and tries to get on with his day. But his wife notices the grimace on his face and instantly recognises that this is definitely something different. She eventually convinces him that this was not something he should dismiss, particularly given that he had the Moderna Spikevax COVID-19 vaccine only 7 days ago. She calls an ambulance and John is taken to a small regional hospital. The nurse seeing him at triage instantly recognises the concern and a troponin level is ordered. This comes back highly elevated and the treating doctor diligently reports this to the Therapeutic Goods Administration (TGA) as a potential case of vaccine-associated myocarditis. Because John needs cardiac monitoring, he is transferred to a larger suburban hospital in the nearest city. Once there he undergoes an angiogram, where they identify a moderate-sized blockage in one of the vessels of his heart. The blockage is from a cholesterol plaque, which has likely been there for many years, but has slowly grown and eventually ruptured to cause a heart attack. The cardiologist places a stent in the artery to open up the blockage and John is quickly discharged so he can return home to recuperate. The report to the TGA remains, unedited, unamended, unaware of all that has happened since.
|
| 6 |
+
|
| 7 |
+
Now contrast this with Lisa. Lisa is an accountant at a large bank. Many years ago, when she was working her way up through the corporate ladder, she had what she calls “the forgotten year”. It was a year of feeling constantly tired, noticing weird rashes, debilitating joint pains and endless trips to many doctors before she was eventually diagnosed with systemic lupus erythematosus (SLE), an autoimmune condition that can affect almost any part of the body. Pham tried a few different medications, before she was eventually enrolled in a clinical trial. This new medication was novel and experimental at the time, but it seemed to work better than the others. In fact, it worked so well that the trial was successful and the drug was eventually approved by the TGA. Fast forward 8 years, Lisa’s SLE has remained in remission. She has been in good health, although she has started noticing a tremor in her hand. Eventually, the tremor gets worse and she sees her GP, who orders some tests and refers her to a neurologist. She is eventually diagnosed with a movement disorder, a broad category of disorders, including Parkinson’s disease. Nobody reports this to the TGA, no known link exists and she has been on this medication for 8 years. But there are 100s of people around the world slowly developing these symptoms, all in isolation with nobody to connect the dots. Eventually, this signal is detected and a warning is introduced. After many years, a gene is identified that predisposes to this risk. But 100s more cases occur before this is known and testing is implemented.
|
| 8 |
+
|
| 9 |
+
What both of these (fictional) cases illustrate is a failure. Not a failure of individuals, but a failure of a system. Pharmacovigilance is the “science and activities relating to the detection, assessment, understanding and prevention of adverse effects or any other medicine/vaccine related problem.” [1] Whilst pharmacovigilance is a necessary part of the drug development phase, many safety signals are too rare (less than one in one thousand) or too delayed to capture in a clinical trials — and this is becoming even more critical as provisional approvals increase, meaning drugs being released with less data and for a smaller patient population where every scrap of data counts (see figure below on provisional approvals). So we must turn instead to what happens after a drug goes to market and identify these safety signals when it is already in use. For such a critical component in the safe development of new and novel medications, one would expect a sophisticated, well-resourced infrastructure that leverages all the advantages of our digital health systems. What we see instead is nothing. Fragmented systems and a lack of vision has lead to a complete void.
|
| 10 |
+
|
| 11 |
+

|
| 12 |
+
|
| 13 |
+
## The Current Landscape
|
| 14 |
+
|
| 15 |
+
The Therapeutic Goods Administration (TGA) in Australia collates post-marketing adverse drug event reports in a central database. This is the Rome of pharmacovigilance – all roads must lead here. But what about the roads? What if I told you there are no roads? Everyone must beat their own path to Rome. All reports to the TGA are human-generated, ad-hoc and one-way. An individual reports to the TGA when they feel it is appropriate, with whatever information they have in hand, with no method to update, refine, revise or review. Some of these reports are highly vetted by expert hospital pharmacovigilance committees, some might be completely fraudulent. None are linked with hospital electronic medical records, Pharmaceutical Benefits Scheme (PBS) prescription data or GP practice management software. Even if they were, none of these systems are linked to eachother, so that a case like John’s would have no information from his subsequent angiogram at another hospital. Good luck finding your way to Rome.
|
| 16 |
+
|
| 17 |
+
The failings in pharmacovigilance reflect the broader failings in digital health. Australia boasts a world-class public health system, with the PBS at the centre of a unified medication landscape. But this unified funding has failed to yield unified digital health systems. My Health Record is an antiquated attempt to bridge these systems. It is a highly incomplete central record of some aspects of healthcare. On an individual level, it sometimes contains some of the information necessary for safe, effective patient care. On a broader level, it provides none of the infrastructure necessary to perform the large scale research needed to improve our knowledge in areas like medication safety and COVID-19 epidemiology.
|
| 18 |
+
|
| 19 |
+
> I just got a foot ultrasound, asked for a copy of the report and if it would be uploaded to [@MyHealthRec](https://twitter.com/MyHealthRec?ref_src=twsrc%5Etfw)
|
| 20 |
+
> Reply: it is company policy to NOT share the results with the patient nor upload it to My health Record 😱 [@AuDigitalHealth](https://twitter.com/AuDigitalHealth?ref_src=twsrc%5Etfw) [@TheInstituteDH](https://twitter.com/TheInstituteDH?ref_src=twsrc%5Etfw)
|
| 21 |
+
>
|
| 22 |
+
> — Daniel Capurro 💉💉💉 (@dcapurro) [February 23, 2021](https://twitter.com/dcapurro/status/1364010140991516676?ref_src=twsrc%5Etfw)
|
| 23 |
+
|
| 24 |
+
## What the Future Might Look Like
|
| 25 |
+
|
| 26 |
+
The US Food and Drugs Administration (FDA) has recently begun grappling with the failed infrastructure of pharmacovigilance. The FDA Sentintel System was developed as a way to integrate insurance databases into their pharmacovigilance program. More recently, the technical shortcomings of insurance databases has lead to investment in a new approach, integrating these insurance databases with large electronic health records (EHRs) in a first step towards building something resembling an integrated digital infrastructure [2]. It should be a great source of shame that Australia, with a mostly public health system, is lagging so far behind our privatised and fragmented cousins in the north.
|
| 27 |
+
|
| 28 |
+
The first steps towards a digital infrastructure that works for better healthcare is data linkage. Adverse drug events are inherently longitudinal, requiring the full breadth of data to be linked across various systems, so that the identifcation of an adverse drug event can be correlated with the initiation of a medication in hospital, or the first script recorded on the PBS. The PBS itself is a notorious black hole for data, taking in the majority of Australia’s prescription medicine data and linking none of it to healthcare records. Contrast this with the proposed Sentinel System, where many private insurance databases will link insurance-funded script data with private EHR data. The ability to link data across jurisdictions, systems and corporate structures must make us reflect on the failings in our own system, which ought to have fewer barriers to overcome.
|
| 29 |
+
|
| 30 |
+
Linked data is only the start, and we will need to invest in developing the sophisticated tooling to take advantage of this. But at least in this aspect we possess the expertise and desire to fill the gap, to modernise pharmacovigilance and meet the needs of the current and future landscape of drug development.
|
| 31 |
+
|
| 32 |
+
[1] Pharmacovigilance, [https://www.who.int/teams/regulation-prequalification/regulation-and-safety/pharmacovigilance](https://www.who.int/teams/regulation-prequalification/regulation-and-safety/pharmacovigilance).
|
| 33 |
+
|
| 34 |
+
[2] Desai RJ, Matheny ME, Johnson K, et al. Broadening the reach of the FDA sentinel system: A roadmap for integrating electronic health record data in a causal analysis framework. _NPJ digital medicine_ 2021; 4: 170.
|
posts/2022-01-29-machine-learning-to-get-stuff-done.md
ADDED
|
@@ -0,0 +1,81 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
Machine Learning To Get Stuff Done
|
| 2 |
+
|
| 3 |
+
I tend to think that there are three categories of machine learning problems. Sometimes machine learning can be really expansive, using sophisticated tools to forge new ground in science. Sometimes it can be incredibly powerful, building large-scale models to perform complex tasks. And sometimes, it can just be the most efficient way to get things done. This is an example of that last category.
|
| 4 |
+
|
| 5 |
+
### Problem
|
| 6 |
+
|
| 7 |
+
I have a project that falls into categories 1 & 2, but in order to do that I need data. Specifically, I need PET-CT scans with features of polymyalgia rheumatica (PMR). But how would I find these? Nobody records this data, it doesn’t exist in a structured, searchable form. But we do have reports — these might technically be searchable, but I don’t think I can easily devise a search term that will capture all the different ways that PMR can be described on a PET-CT report (including many instances where some classic features are described but the words “polymyalgia rheumatica” never appear in the report).
|
| 8 |
+
|
| 9 |
+
### Solution
|
| 10 |
+
|
| 11 |
+
I chose to tackle this as a machine learning problem. Specifically, a supervised learning problem. But to do this I needed data. The first step is always to collect data.
|
| 12 |
+
|
| 13 |
+
### Data Collection
|
| 14 |
+
|
| 15 |
+
I needed a collection of PET-CT reports and I needed labels for this reports. I originally thought that I would label words and phrases within the reports to do _named entity recognition_ , a task that involves training a model to identify these features in text. But there are many potential PMR features noted in PET-CT reports, making this a fairly time-consuming process. Instead I settled on simply classifying the reports as either **PMR** or **Not PMR**.
|
| 16 |
+
|
| 17 |
+
To generate this data, I turned to my data annotation tool [prodigy](prodi.gy). I wrote a custom data loader using an SQL query and some python code to trim the reports so they just contained the reporting physicians notes and conclusion — this last step is necessary because the text of the original request can bias the model and ultimately doesn’t contain information about the PET-CT itself. This code pulls PET-CT reports directly from our research data warehouse one at a time, so no intermediate step is required to generate a large CSV file or spreadsheet. You can try a demo of prodigy [here](https://prodi.gy/demo) — it has a clean, intuitive interface and is a breeze to customize for specific tasks.
|
| 18 |
+
|
| 19 |
+
An example of the prodigy interface.
|
| 20 |
+
|
| 21 |
+
### Training an Initial Model
|
| 22 |
+
|
| 23 |
+
I originally trained a quick model in R to obtain a baseline and confirm that this was all feasible. For this I generated skip-grams (n=3, k=1) and performed **term frequency–inverse document frequency (TF-IDF)** to obtain the features. The very quick explanation of this is that I pulled out single words and clusters of up to three words (the skip part means that those words do not have to be adjacent, but may be separated by 1 word), I then counted how many times these skip grams occurred in each report and normalised this by the frequency across all reports (so that infrequent skip grams carry more value). I then trained a XGBoost model on these features to classify reports as being either **PMR** or **Not PMR**.
|
| 24 |
+
|
| 25 |
+
```
|
| 26 |
+
library(tidyverse)
|
| 27 |
+
library(tidymodels)
|
| 28 |
+
library(textrecipes)
|
| 29 |
+
library(fastshap)
|
| 30 |
+
|
| 31 |
+
# Split the data into trian and test
|
| 32 |
+
split <- initial_split(data = data, strata = pmr, prop = .8)
|
| 33 |
+
train <- training(split)
|
| 34 |
+
test <- testing(split)
|
| 35 |
+
|
| 36 |
+
# Specify the recipe that creates skip grams, retains only the 1000 most common skip grams, then performs TF-IDF
|
| 37 |
+
rec <- recipe(pmr ~ text, data = train) %>%
|
| 38 |
+
step_tokenize(text, token = "skip_ngrams", options = list(n_min = 1, n = 3, k = 1)) %>%
|
| 39 |
+
step_tokenfilter(text, max_tokens = 1000) %>%
|
| 40 |
+
step_tfidf(text)
|
| 41 |
+
|
| 42 |
+
# Specify the xgboost model with some hyperparameters
|
| 43 |
+
model <- boost_tree(trees = 1000,
|
| 44 |
+
mtry = 10,
|
| 45 |
+
min_n = 3,
|
| 46 |
+
tree_depth = 5) %>%
|
| 47 |
+
set_engine("xgboost", eval_metric = 'error') %>%
|
| 48 |
+
set_mode("classification")
|
| 49 |
+
|
| 50 |
+
## Connect the recipe and model in a workflow
|
| 51 |
+
wf <- workflow() %>%
|
| 52 |
+
add_recipe(rec) %>%
|
| 53 |
+
add_model(model) %>%
|
| 54 |
+
fit(train)
|
| 55 |
+
|
| 56 |
+
## Specify the metrics we would like to report
|
| 57 |
+
model_metrics <- metric_set(roc_auc, pr_auc, accuracy)
|
| 58 |
+
|
| 59 |
+
## Trian the model on the train set and test on test set (as specified by split)
|
| 60 |
+
res <- last_fit(wf, split, metrics = model_metrics)
|
| 61 |
+
```
|
| 62 |
+
|
| 63 |
+
This model has an AUC of about 0.95 and an accuracy of just over 90% — a pretty good start! Examining the Shapley values (a way of estimating the contribution of each skip gram to the model prediction), it seems to be using some of those PMR features (particularly ischial tuberosities), but there probably some spurious features here. My feeling is that this model will probably suffer from over-fitting.
|
| 64 |
+
|
| 65 |
+
Shapley values from the XGBoost model.
|
| 66 |
+
|
| 67 |
+
### Training a Transformers Model
|
| 68 |
+
|
| 69 |
+
This is when I moved back to python and the Hugging Face 🤗 [Transformers library](https://huggingface.co/docs/transformers/index). I have previously pre-trained a model on a large number of documents from our hospital, which I then used for [Adverse Drug Reaction detection](https://www.medrxiv.org/content/10.1101/2021.12.11.21267504v2). Since then, I have updated this pre-trained model, training on well over 1 million documents (discharge summaries, radiology reports, pathology reports, progress notes). This means that we can start with a model that has a good idea of the general structure and content of our documents, including medical imaging reports.
|
| 70 |
+
|
| 71 |
+
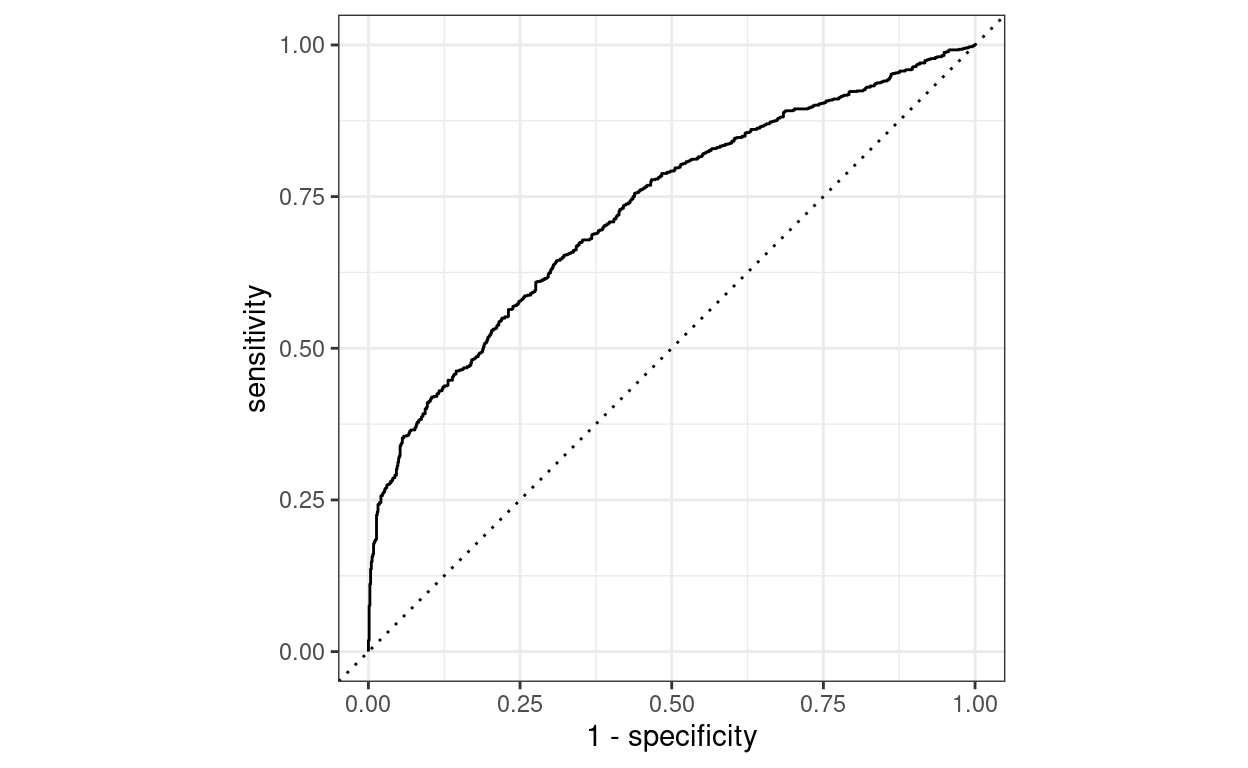
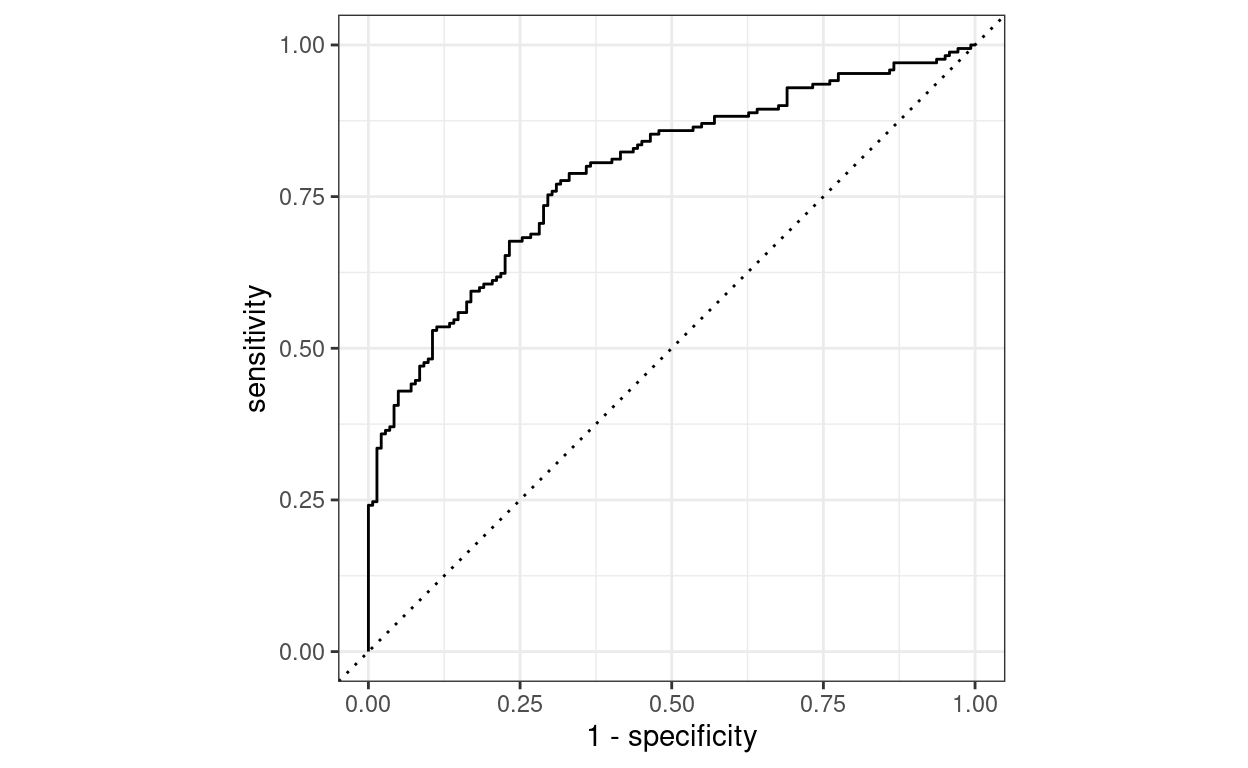
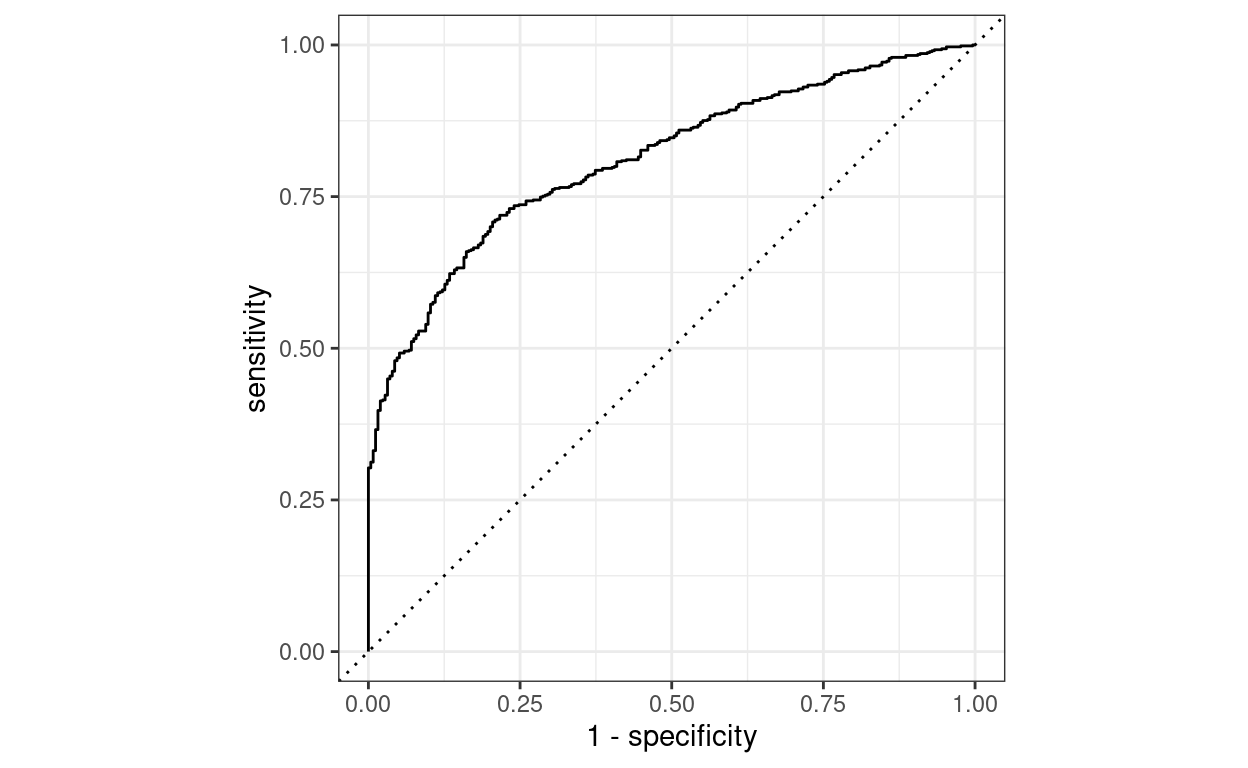
I fine-tuned this model on my small dataset of labelled PET-CT reports, which took less than 1 minute on 3x 1080ti GPUs that we have running in our department. The final ROC curve looks pretty good!
|
| 72 |
+
|
| 73 |
+

|
| 74 |
+
|
| 75 |
+
### Reading 38,000 Reports
|
| 76 |
+
|
| 77 |
+
Finally, I was able to pull every PET-CT report from our research data warehouse and run the model across them to see how many contained PMR features. That’s where I got the 305 number that I tweeted about.
|
| 78 |
+
|
| 79 |
+
> How many of the 38,000 PET scan reports at my hospital were reported to have features of polymyalgia rheumatica? 305. Did I read all 38,000 to find this out? Hell no, I built a model to do it for me.
|
| 80 |
+
>
|
| 81 |
+
> — Christopher McMaster (@DrCMcMaster) [January 27, 2022](https://twitter.com/DrCMcMaster/status/1486592038862655498?ref_src=twsrc%5Etfw)
|
posts/images/BPE.drawio.svg
ADDED

|
posts/images/Recurrent_neural_network_unfold.svg
ADDED

|
posts/images/paste-35A12A44.png
ADDED

|
posts/images/prodigy.svg
ADDED

|
posts/images/unnamed-chunk-1-1.png
ADDED

|
posts/images/unnamed-chunk-11-1.png
ADDED
|
posts/images/unnamed-chunk-15-1.png
ADDED

|
posts/images/unnamed-chunk-16-1.png
ADDED
|
posts/images/unnamed-chunk-7-1.png
ADDED

|
posts/images/unnamed-chunk-9-1.png
ADDED
|
requirements.txt
ADDED
|
@@ -0,0 +1,4 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
python-fasthtml
|
| 2 |
+
starlette
|
| 3 |
+
numpy
|
| 4 |
+
scikit-learn
|
static/css/style.css
ADDED
|
@@ -0,0 +1,195 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
@import url('https://fonts.googleapis.com/css2?family=Poppins:wght@300;400;600&display=swap');
|
| 2 |
+
|
| 3 |
+
body {
|
| 4 |
+
font-family: Charter, "Bitstream Charter", "Sitka Text", Cambria, serif;
|
| 5 |
+
line-height: 1.6;
|
| 6 |
+
margin: 0 auto;
|
| 7 |
+
max-width: 35em;
|
| 8 |
+
padding: 1em;
|
| 9 |
+
}
|
| 10 |
+
|
| 11 |
+
h1, h2, h3, h4, h5, h6 {
|
| 12 |
+
font-family: Charter, "Bitstream Charter", "Sitka Text", Cambria, serif;
|
| 13 |
+
font-weight: normal;
|
| 14 |
+
}
|
| 15 |
+
|
| 16 |
+
a {
|
| 17 |
+
color: currentColor;
|
| 18 |
+
text-decoration-thickness: 1px;
|
| 19 |
+
text-underline-offset: 0.2em;
|
| 20 |
+
}
|
| 21 |
+
|
| 22 |
+
nav {
|
| 23 |
+
margin: 2em 0 4em 0;
|
| 24 |
+
padding-bottom: 0.5em;
|
| 25 |
+
border-bottom: 1px solid #ddd;
|
| 26 |
+
}
|
| 27 |
+
|
| 28 |
+
nav a {
|
| 29 |
+
margin-right: 2em;
|
| 30 |
+
text-decoration: none;
|
| 31 |
+
}
|
| 32 |
+
|
| 33 |
+
nav a:hover {
|
| 34 |
+
text-decoration: underline;
|
| 35 |
+
}
|
| 36 |
+
|
| 37 |
+
nav a.active {
|
| 38 |
+
text-decoration: underline;
|
| 39 |
+
text-decoration-thickness: 2px;
|
| 40 |
+
}
|
| 41 |
+
|
| 42 |
+
img {
|
| 43 |
+
max-width: 100%;
|
| 44 |
+
height: auto;
|
| 45 |
+
}
|
| 46 |
+
|
| 47 |
+
.profile {
|
| 48 |
+
margin: 3em 0;
|
| 49 |
+
}
|
| 50 |
+
|
| 51 |
+
.profile img {
|
| 52 |
+
width: 100px;
|
| 53 |
+
height: 100px;
|
| 54 |
+
}
|
| 55 |
+
|
| 56 |
+
.date {
|
| 57 |
+
font-size: 0.9em;
|
| 58 |
+
}
|
| 59 |
+
|
| 60 |
+
.blog-post {
|
| 61 |
+
margin: 2em 0;
|
| 62 |
+
}
|
| 63 |
+
|
| 64 |
+
.button {
|
| 65 |
+
display: inline-block;
|
| 66 |
+
padding: 0.5em 1em;
|
| 67 |
+
border: 1px solid currentColor;
|
| 68 |
+
text-decoration: none;
|
| 69 |
+
}
|
| 70 |
+
|
| 71 |
+
.button:hover {
|
| 72 |
+
background: black;
|
| 73 |
+
color: white;
|
| 74 |
+
border-color: black;
|
| 75 |
+
}
|
| 76 |
+
|
| 77 |
+
/* Add subtle transitions */
|
| 78 |
+
a, .button {
|
| 79 |
+
transition: all 0.2s ease;
|
| 80 |
+
}
|
| 81 |
+
|
| 82 |
+
.similar-button-container {
|
| 83 |
+
margin-top: 2rem;
|
| 84 |
+
margin-bottom: 2rem;
|
| 85 |
+
text-align: center;
|
| 86 |
+
}
|
| 87 |
+
|
| 88 |
+
.similar-button-container .button {
|
| 89 |
+
padding: 0.8rem 1.5rem;
|
| 90 |
+
background-color: #4a5568;
|
| 91 |
+
color: white;
|
| 92 |
+
border: none;
|
| 93 |
+
border-radius: 4px;
|
| 94 |
+
cursor: pointer;
|
| 95 |
+
font-size: 1.1em;
|
| 96 |
+
}
|
| 97 |
+
|
| 98 |
+
.similar-button-container .button:hover {
|
| 99 |
+
background-color: #2d3748;
|
| 100 |
+
transform: translateY(-1px);
|
| 101 |
+
transition: all 0.2s ease;
|
| 102 |
+
}
|
| 103 |
+
|
| 104 |
+
.abstract-result {
|
| 105 |
+
margin-bottom: 2rem;
|
| 106 |
+
padding-bottom: 1rem;
|
| 107 |
+
border-bottom: 1px solid #eee;
|
| 108 |
+
}
|
| 109 |
+
|
| 110 |
+
.similar-button {
|
| 111 |
+
font-size: 0.9em;
|
| 112 |
+
padding: 0.3rem 0.8rem;
|
| 113 |
+
margin-top: 0.5rem;
|
| 114 |
+
background-color: #4a5568;
|
| 115 |
+
color: white;
|
| 116 |
+
border: none;
|
| 117 |
+
border-radius: 4px;
|
| 118 |
+
cursor: pointer;
|
| 119 |
+
}
|
| 120 |
+
|
| 121 |
+
.similar-button:hover {
|
| 122 |
+
background-color: #2d3748;
|
| 123 |
+
}
|
| 124 |
+
|
| 125 |
+
.similar-results {
|
| 126 |
+
margin-top: 1rem;
|
| 127 |
+
padding-left: 1rem;
|
| 128 |
+
border-left: 3px solid #eee;
|
| 129 |
+
}
|
| 130 |
+
|
| 131 |
+
.modal-overlay {
|
| 132 |
+
display: none;
|
| 133 |
+
position: fixed;
|
| 134 |
+
top: 0;
|
| 135 |
+
left: 0;
|
| 136 |
+
width: 100%;
|
| 137 |
+
height: 100%;
|
| 138 |
+
background-color: rgba(0, 0, 0, 0.75);
|
| 139 |
+
z-index: 1000;
|
| 140 |
+
}
|
| 141 |
+
|
| 142 |
+
.modal {
|
| 143 |
+
position: fixed;
|
| 144 |
+
top: 50%;
|
| 145 |
+
left: 50%;
|
| 146 |
+
transform: translate(-50%, -50%);
|
| 147 |
+
background-color: #1a1a1a;
|
| 148 |
+
color: #ffffff;
|
| 149 |
+
padding: 2rem;
|
| 150 |
+
border-radius: 8px;
|
| 151 |
+
max-width: 80%;
|
| 152 |
+
max-height: 80vh;
|
| 153 |
+
overflow-y: auto;
|
| 154 |
+
z-index: 1001;
|
| 155 |
+
box-shadow: 0 0 20px rgba(0, 0, 0, 0.5);
|
| 156 |
+
}
|
| 157 |
+
|
| 158 |
+
.modal-close {
|
| 159 |
+
position: absolute;
|
| 160 |
+
top: 1rem;
|
| 161 |
+
right: 1rem;
|
| 162 |
+
font-size: 1.5rem;
|
| 163 |
+
cursor: pointer;
|
| 164 |
+
border: none;
|
| 165 |
+
background: none;
|
| 166 |
+
color: #ffffff;
|
| 167 |
+
padding: 0.5rem;
|
| 168 |
+
}
|
| 169 |
+
|
| 170 |
+
.modal-close:hover {
|
| 171 |
+
color: #cccccc;
|
| 172 |
+
}
|
| 173 |
+
|
| 174 |
+
.modal a {
|
| 175 |
+
color: #66b3ff;
|
| 176 |
+
}
|
| 177 |
+
|
| 178 |
+
.modal a:hover {
|
| 179 |
+
color: #99ccff;
|
| 180 |
+
}
|
| 181 |
+
|
| 182 |
+
.similar-list {
|
| 183 |
+
list-style: none;
|
| 184 |
+
padding: 0;
|
| 185 |
+
}
|
| 186 |
+
|
| 187 |
+
.similar-abstract {
|
| 188 |
+
margin-bottom: 2rem;
|
| 189 |
+
padding-bottom: 1rem;
|
| 190 |
+
border-bottom: 1px solid #333;
|
| 191 |
+
}
|
| 192 |
+
|
| 193 |
+
.similar-abstract:last-child {
|
| 194 |
+
border-bottom: none;
|
| 195 |
+
}
|